# A tibble: 22 × 3
id job price_glass
<int> <chr> <chr>
1 1 Student 0
2 2 Retired 0
3 3 Other 0
4 4 Employed 10
5 5 Employed See comment
6 6 Student 05-Oct
# … with 16 more rowsrbtl - Data wrangling with tidyr
Lars Schöbitz
Global Health Engineering - ETH Zurich
2022-05-19
Today
- Part 1: Data types and vectors
- Live Coding Exercise
- Part 2: tidyr - long and wide formats
- Live Coding Exercise
- Part 3: dplyr - joining data
- Live Coding Exercise
- Homework Assignment 13
- Programming Exercise
Learning Objectives
- Learners can apply functions from the
tidyr(actually dplyr) R Package to join multiple data sets - Learners can apply functions from the tidyr R Package to transform their data from a wide to a long format and vice versa
Part 1: Data types and vectors
Why care about data types?
Example: survey data
Oh why won’t you work?!
Oh why won’t you still work??!!
Take a breath and look at your data
Very common data tidying step!
Very common data tidying step!
# A tibble: 22 × 4
id job price_glass_new price_glass
<int> <chr> <chr> <chr>
1 1 Student 0 0
2 2 Retired 0 0
3 3 Other 0 0
4 4 Employed 10 10
5 5 Employed <NA> See comment
6 6 Student 7.5 05-Oct
7 7 Student 0 0
8 8 Retired 0 0
9 9 Student 10 10
10 10 Employed 0 0
11 11 Employed 20 20 (2chf per person with 10 pe…
12 12 Student 10 10
13 13 Student 10 10
14 14 Employed 0 0
15 15 Student 10 10
16 16 Student 0 0
17 17 Employed 7.5 5 to 10
18 18 Other 0 0
19 19 Student 0 0
20 20 Employed 10 10
21 21 Employed 0 0
22 22 Employed 5 5 Sumamrise? Argh!!!!
survey_data_small %>%
mutate(price_glass_new = case_when(
price_glass == "5 to 10" ~ "7.5",
price_glass == "05-Oct" ~ "7.5",
str_detect(price_glass, pattern = "20") == TRUE ~ "20",
str_detect(price_glass, pattern = "See comment") == TRUE ~ NA_character_,
TRUE ~ price_glass
)) %>%
summarise(mean_price_glass = mean(price_glass_new, na.rm = TRUE))# A tibble: 1 × 1
mean_price_glass
<dbl>
1 NAAlways respect your data types!
survey_data_small %>%
mutate(price_glass_new = case_when(
price_glass == "5 to 10" ~ "7.5",
price_glass == "05-Oct" ~ "7.5",
str_detect(price_glass, pattern = "20") == TRUE ~ "20",
str_detect(price_glass, pattern = "See comment") == TRUE ~ NA_character_,
TRUE ~ price_glass
)) %>%
mutate(price_glass_new = as.numeric(price_glass_new)) %>%
summarise(mean_price_glass = mean(price_glass_new, na.rm = TRUE))# A tibble: 1 × 1
mean_price_glass
<dbl>
1 4.76Live Coding Exercise
ae-13-data-wrangling-tidyr
- Head over to the GitHub Organisation for the course.
- Find the repo for week 13 that has your GitHub username.
- Clone the repo with your username to the RStudio Cloud.
- Open the file:
ae-13a-tidyr.qmd - Use your Sticky Notes to let me know when you are ready.
Break One

15:00
Part 2: tidyr - long and wide formats
.
.
.
A grammar of data tidying

The goal of tidyr is to help you tidy your data via
- pivoting for going between wide and long data
- splitting and combining character columns
- nesting and unnesting columns
- clarifying how
NAs should be treated
Pivoting data
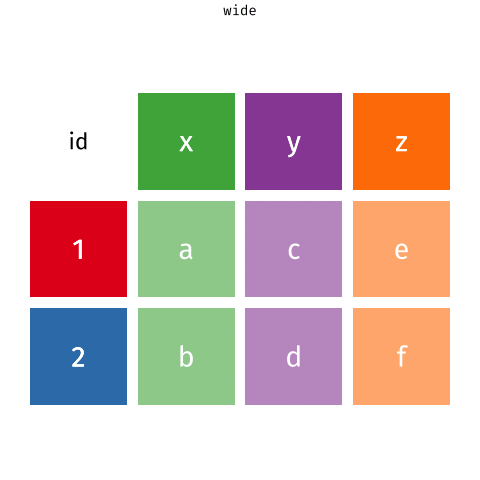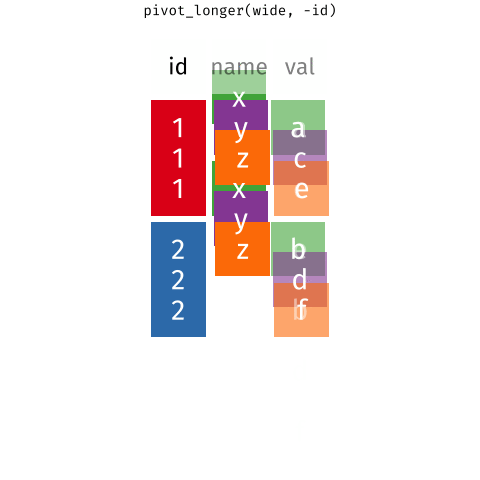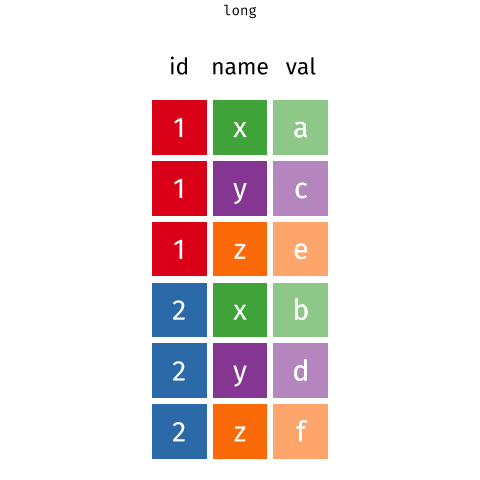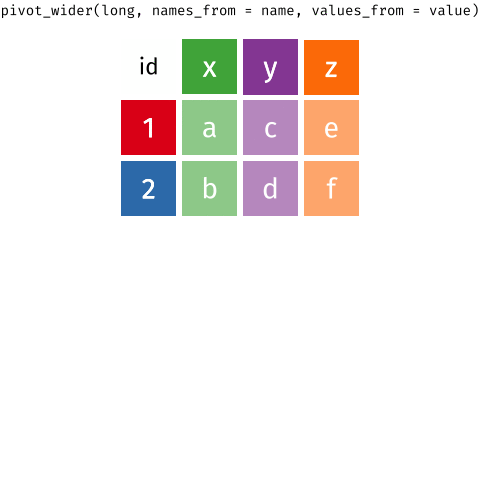
Waste characterisation data
| objid | location | pet | metal_alu | glass | paper | recyclable | non_recyclable | total |
|---|---|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 0.92 | 1.14 | 2.05 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 0.61 | 3.04 | 3.64 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 0.41 | 1.57 | 1.99 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 1.56 | 0.62 | 2.19 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.54 | 0.58 | 1.12 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.16 | 0.34 | 0.50 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 0.81 | 1.52 | 2.33 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 2.08 | 1.99 | 4.07 |
How would you plot this?
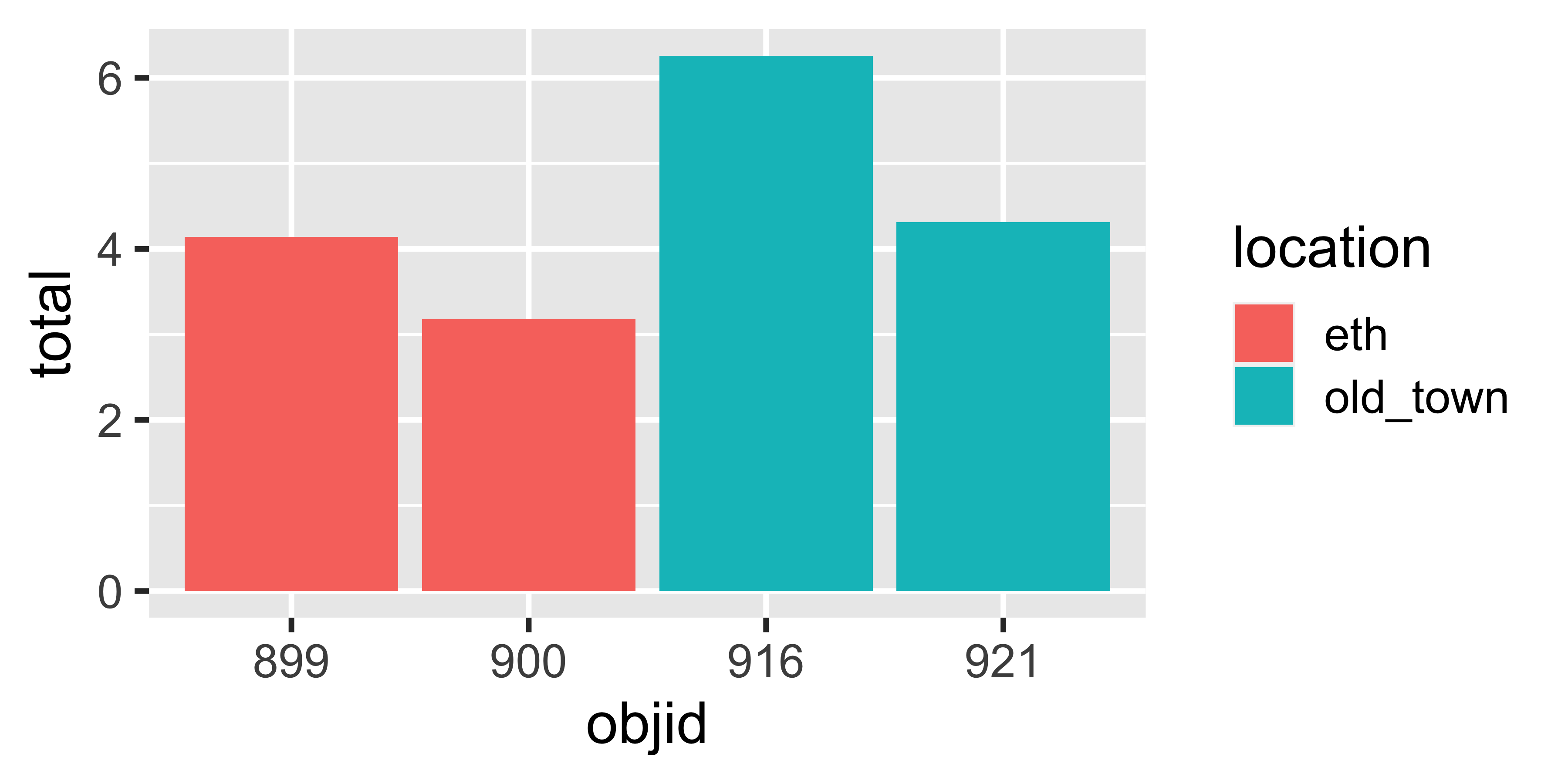
And this?

You need: A long format
| objid | location | waste_category | weight |
|---|---|---|---|
| 900 | eth | pet | 0.06 |
| 900 | eth | metal_alu | 0.06 |
| 900 | eth | glass | 0.58 |
| 900 | eth | paper | 0.21 |
| 900 | eth | other | 1.14 |
| 899 | eth | pet | 0.14 |
| 899 | eth | metal_alu | 0.01 |
| 899 | eth | glass | 0.18 |
| 899 | eth | paper | 0.28 |
| 899 | eth | other | 3.04 |
| 921 | old_town | pet | 0.00 |
| 921 | old_town | metal_alu | 0.00 |
| 921 | old_town | glass | 0.00 |
| 921 | old_town | paper | 0.41 |
| 921 | old_town | other | 1.57 |
| 916 | old_town | pet | 0.17 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.80 |
| 916 | old_town | paper | 0.55 |
| 916 | old_town | other | 0.62 |
| 900 | eth | pet | 0.10 |
| 900 | eth | metal_alu | 0.04 |
| 900 | eth | glass | 0.00 |
| 900 | eth | paper | 0.40 |
| 900 | eth | other | 0.58 |
| 899 | eth | pet | 0.08 |
| 899 | eth | metal_alu | 0.03 |
| 899 | eth | glass | 0.00 |
| 899 | eth | paper | 0.05 |
| 899 | eth | other | 0.34 |
| 921 | old_town | pet | 0.08 |
| 921 | old_town | metal_alu | 0.03 |
| 921 | old_town | glass | 0.30 |
| 921 | old_town | paper | 0.40 |
| 921 | old_town | other | 1.52 |
| 916 | old_town | pet | 0.11 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.92 |
| 916 | old_town | paper | 1.01 |
| 916 | old_town | other | 1.99 |
Reminder: The wide format
| objid | location | pet | metal_alu | glass | paper | recyclable | non_recyclable | total |
|---|---|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 0.92 | 1.14 | 2.05 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 0.61 | 3.04 | 3.64 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 0.41 | 1.57 | 1.99 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 1.56 | 0.62 | 2.19 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.54 | 0.58 | 1.12 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.16 | 0.34 | 0.50 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 0.81 | 1.52 | 2.33 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 2.08 | 1.99 | 4.07 |
Or this?
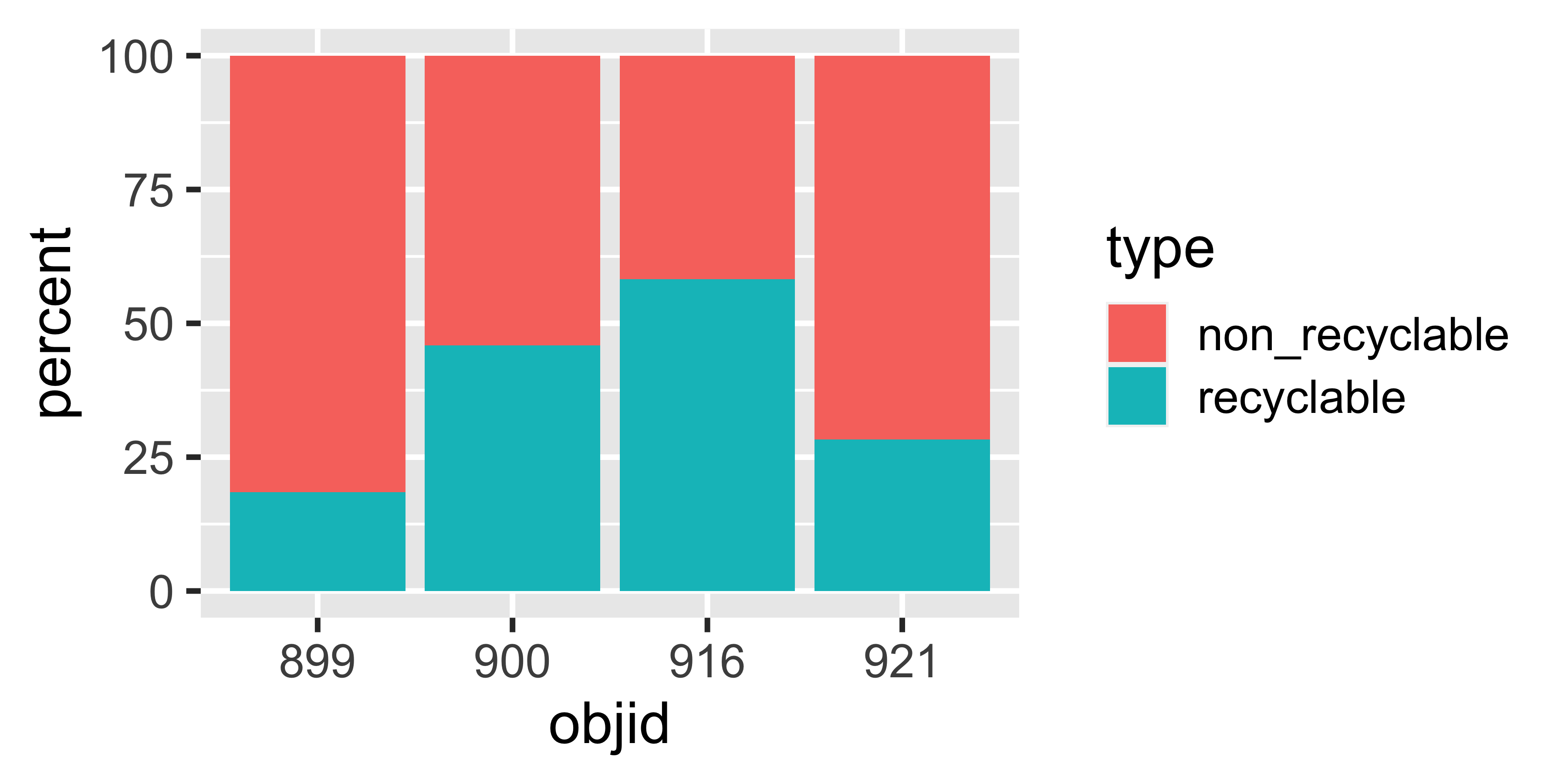
Calculate percentages
| objid | location | waste_category | type | weight | percent |
|---|---|---|---|---|---|
| 900 | eth | pet | recyclable | 0.06 | 2.02 |
| 900 | eth | metal_alu | recyclable | 0.06 | 1.95 |
| 900 | eth | glass | recyclable | 0.58 | 18.14 |
| 900 | eth | paper | recyclable | 0.21 | 6.74 |
| 900 | eth | other | non_recyclable | 1.14 | 35.78 |
| 899 | eth | pet | recyclable | 0.14 | 3.33 |
| 899 | eth | metal_alu | recyclable | 0.01 | 0.31 |
| 899 | eth | glass | recyclable | 0.18 | 4.30 |
| 899 | eth | paper | recyclable | 0.28 | 6.69 |
| 899 | eth | other | non_recyclable | 3.04 | 73.36 |
| 921 | old_town | pet | recyclable | 0.00 | 0.00 |
| 921 | old_town | metal_alu | recyclable | 0.00 | 0.00 |
| 921 | old_town | glass | recyclable | 0.00 | 0.00 |
| 921 | old_town | paper | recyclable | 0.41 | 9.60 |
| 921 | old_town | other | non_recyclable | 1.57 | 36.46 |
| 916 | old_town | pet | recyclable | 0.17 | 2.76 |
| 916 | old_town | metal_alu | recyclable | 0.04 | 0.69 |
| 916 | old_town | glass | recyclable | 0.80 | 12.73 |
| 916 | old_town | paper | recyclable | 0.55 | 8.82 |
| 916 | old_town | other | non_recyclable | 0.62 | 9.99 |
| 900 | eth | pet | recyclable | 0.10 | 3.09 |
| 900 | eth | metal_alu | recyclable | 0.04 | 1.35 |
| 900 | eth | glass | recyclable | 0.00 | 0.00 |
| 900 | eth | paper | recyclable | 0.40 | 12.60 |
| 900 | eth | other | non_recyclable | 0.58 | 18.33 |
| 899 | eth | pet | recyclable | 0.08 | 1.86 |
| 899 | eth | metal_alu | recyclable | 0.03 | 0.72 |
| 899 | eth | glass | recyclable | 0.00 | 0.00 |
| 899 | eth | paper | recyclable | 0.05 | 1.26 |
| 899 | eth | other | non_recyclable | 0.34 | 8.16 |
| 921 | old_town | pet | recyclable | 0.08 | 1.81 |
| 921 | old_town | metal_alu | recyclable | 0.03 | 0.70 |
| 921 | old_town | glass | recyclable | 0.30 | 6.89 |
| 921 | old_town | paper | recyclable | 0.40 | 9.32 |
| 921 | old_town | other | non_recyclable | 1.52 | 35.21 |
| 916 | old_town | pet | recyclable | 0.11 | 1.74 |
| 916 | old_town | metal_alu | recyclable | 0.04 | 0.70 |
| 916 | old_town | glass | recyclable | 0.92 | 14.63 |
| 916 | old_town | paper | recyclable | 1.01 | 16.20 |
| 916 | old_town | other | non_recyclable | 1.99 | 31.73 |
How to
| objid | location | pet | metal_alu | glass | paper | recyclable | non_recyclable | total |
|---|---|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 0.92 | 1.14 | 2.05 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 0.61 | 3.04 | 3.64 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 0.41 | 1.57 | 1.99 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 1.56 | 0.62 | 2.19 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.54 | 0.58 | 1.12 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.16 | 0.34 | 0.50 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 0.81 | 1.52 | 2.33 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 2.08 | 1.99 | 4.07 |
How to
| objid | location | pet | metal_alu | glass | paper | non_recyclable |
|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 1.14 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 3.04 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 1.57 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 0.62 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.58 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.34 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 1.52 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 1.99 |
How to
| objid | location | pet | metal_alu | glass | paper | other |
|---|---|---|---|---|---|---|
| 900 | eth | 0.06 | 0.06 | 0.58 | 0.21 | 1.14 |
| 899 | eth | 0.14 | 0.01 | 0.18 | 0.28 | 3.04 |
| 921 | old_town | 0.00 | 0.00 | 0.00 | 0.41 | 1.57 |
| 916 | old_town | 0.17 | 0.04 | 0.80 | 0.55 | 0.62 |
| 900 | eth | 0.10 | 0.04 | 0.00 | 0.40 | 0.58 |
| 899 | eth | 0.08 | 0.03 | 0.00 | 0.05 | 0.34 |
| 921 | old_town | 0.08 | 0.03 | 0.30 | 0.40 | 1.52 |
| 916 | old_town | 0.11 | 0.04 | 0.92 | 1.01 | 1.99 |
How to
waste_category_levels <- c("glass", "metal_alu", "paper", "pet", "other")
waste_data_untidy %>%
select(objid:paper, non_recyclable) %>%
rename(other = non_recyclable) %>%
pivot_longer(cols = pet:other,
names_to = "waste_category",
values_to = "weight") %>%
mutate(waste_category = factor(waste_category,
levels = waste_category_levels)) | objid | location | waste_category | weight |
|---|---|---|---|
| 900 | eth | pet | 0.06 |
| 900 | eth | metal_alu | 0.06 |
| 900 | eth | glass | 0.58 |
| 900 | eth | paper | 0.21 |
| 900 | eth | other | 1.14 |
| 899 | eth | pet | 0.14 |
| 899 | eth | metal_alu | 0.01 |
| 899 | eth | glass | 0.18 |
| 899 | eth | paper | 0.28 |
| 899 | eth | other | 3.04 |
| 921 | old_town | pet | 0.00 |
| 921 | old_town | metal_alu | 0.00 |
| 921 | old_town | glass | 0.00 |
| 921 | old_town | paper | 0.41 |
| 921 | old_town | other | 1.57 |
| 916 | old_town | pet | 0.17 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.80 |
| 916 | old_town | paper | 0.55 |
| 916 | old_town | other | 0.62 |
| 900 | eth | pet | 0.10 |
| 900 | eth | metal_alu | 0.04 |
| 900 | eth | glass | 0.00 |
| 900 | eth | paper | 0.40 |
| 900 | eth | other | 0.58 |
| 899 | eth | pet | 0.08 |
| 899 | eth | metal_alu | 0.03 |
| 899 | eth | glass | 0.00 |
| 899 | eth | paper | 0.05 |
| 899 | eth | other | 0.34 |
| 921 | old_town | pet | 0.08 |
| 921 | old_town | metal_alu | 0.03 |
| 921 | old_town | glass | 0.30 |
| 921 | old_town | paper | 0.40 |
| 921 | old_town | other | 1.52 |
| 916 | old_town | pet | 0.11 |
| 916 | old_town | metal_alu | 0.04 |
| 916 | old_town | glass | 0.92 |
| 916 | old_town | paper | 1.01 |
| 916 | old_town | other | 1.99 |
How to
waste_category_levels <- c("glass", "metal_alu", "paper", "pet", "other")
waste_data_untidy %>%
select(objid:paper, non_recyclable) %>%
rename(other = non_recyclable) %>%
pivot_longer(cols = pet:other,
names_to = "waste_category",
values_to = "weight") %>%
mutate(waste_category = factor(waste_category,
levels = waste_category_levels)) %>%
mutate(type = case_when(
waste_category == "other" ~ "non_recyclable",
TRUE ~ "recyclable")) %>%
relocate(type, .before = weight)| objid | location | waste_category | type | weight |
|---|---|---|---|---|
| 900 | eth | pet | recyclable | 0.06 |
| 900 | eth | metal_alu | recyclable | 0.06 |
| 900 | eth | glass | recyclable | 0.58 |
| 900 | eth | paper | recyclable | 0.21 |
| 900 | eth | other | non_recyclable | 1.14 |
| 899 | eth | pet | recyclable | 0.14 |
| 899 | eth | metal_alu | recyclable | 0.01 |
| 899 | eth | glass | recyclable | 0.18 |
| 899 | eth | paper | recyclable | 0.28 |
| 899 | eth | other | non_recyclable | 3.04 |
| 921 | old_town | pet | recyclable | 0.00 |
| 921 | old_town | metal_alu | recyclable | 0.00 |
| 921 | old_town | glass | recyclable | 0.00 |
| 921 | old_town | paper | recyclable | 0.41 |
| 921 | old_town | other | non_recyclable | 1.57 |
| 916 | old_town | pet | recyclable | 0.17 |
| 916 | old_town | metal_alu | recyclable | 0.04 |
| 916 | old_town | glass | recyclable | 0.80 |
| 916 | old_town | paper | recyclable | 0.55 |
| 916 | old_town | other | non_recyclable | 0.62 |
| 900 | eth | pet | recyclable | 0.10 |
| 900 | eth | metal_alu | recyclable | 0.04 |
| 900 | eth | glass | recyclable | 0.00 |
| 900 | eth | paper | recyclable | 0.40 |
| 900 | eth | other | non_recyclable | 0.58 |
| 899 | eth | pet | recyclable | 0.08 |
| 899 | eth | metal_alu | recyclable | 0.03 |
| 899 | eth | glass | recyclable | 0.00 |
| 899 | eth | paper | recyclable | 0.05 |
| 899 | eth | other | non_recyclable | 0.34 |
| 921 | old_town | pet | recyclable | 0.08 |
| 921 | old_town | metal_alu | recyclable | 0.03 |
| 921 | old_town | glass | recyclable | 0.30 |
| 921 | old_town | paper | recyclable | 0.40 |
| 921 | old_town | other | non_recyclable | 1.52 |
| 916 | old_town | pet | recyclable | 0.11 |
| 916 | old_town | metal_alu | recyclable | 0.04 |
| 916 | old_town | glass | recyclable | 0.92 |
| 916 | old_town | paper | recyclable | 1.01 |
| 916 | old_town | other | non_recyclable | 1.99 |
How to
waste_category_levels <- c("glass", "metal_alu", "paper", "pet", "other")
waste_data_untidy %>%
select(objid:paper, non_recyclable) %>%
rename(other = non_recyclable) %>%
pivot_longer(cols = pet:other,
names_to = "waste_category",
values_to = "weight") %>%
mutate(waste_category = factor(waste_category,
levels = waste_category_levels)) %>%
mutate(type = case_when(
waste_category == "other" ~ "non_recyclable",
TRUE ~ "recyclable")) %>%
relocate(type, .before = weight) %>%
group_by(objid) %>%
mutate(percent = weight / sum(weight) * 100)| objid | location | waste_category | type | weight | percent |
|---|---|---|---|---|---|
| 900 | eth | pet | recyclable | 0.06 | 2.02 |
| 900 | eth | metal_alu | recyclable | 0.06 | 1.95 |
| 900 | eth | glass | recyclable | 0.58 | 18.14 |
| 900 | eth | paper | recyclable | 0.21 | 6.74 |
| 900 | eth | other | non_recyclable | 1.14 | 35.78 |
| 899 | eth | pet | recyclable | 0.14 | 3.33 |
| 899 | eth | metal_alu | recyclable | 0.01 | 0.31 |
| 899 | eth | glass | recyclable | 0.18 | 4.30 |
| 899 | eth | paper | recyclable | 0.28 | 6.69 |
| 899 | eth | other | non_recyclable | 3.04 | 73.36 |
| 921 | old_town | pet | recyclable | 0.00 | 0.00 |
| 921 | old_town | metal_alu | recyclable | 0.00 | 0.00 |
| 921 | old_town | glass | recyclable | 0.00 | 0.00 |
| 921 | old_town | paper | recyclable | 0.41 | 9.60 |
| 921 | old_town | other | non_recyclable | 1.57 | 36.46 |
| 916 | old_town | pet | recyclable | 0.17 | 2.76 |
| 916 | old_town | metal_alu | recyclable | 0.04 | 0.69 |
| 916 | old_town | glass | recyclable | 0.80 | 12.73 |
| 916 | old_town | paper | recyclable | 0.55 | 8.82 |
| 916 | old_town | other | non_recyclable | 0.62 | 9.99 |
| 900 | eth | pet | recyclable | 0.10 | 3.09 |
| 900 | eth | metal_alu | recyclable | 0.04 | 1.35 |
| 900 | eth | glass | recyclable | 0.00 | 0.00 |
| 900 | eth | paper | recyclable | 0.40 | 12.60 |
| 900 | eth | other | non_recyclable | 0.58 | 18.33 |
| 899 | eth | pet | recyclable | 0.08 | 1.86 |
| 899 | eth | metal_alu | recyclable | 0.03 | 0.72 |
| 899 | eth | glass | recyclable | 0.00 | 0.00 |
| 899 | eth | paper | recyclable | 0.05 | 1.26 |
| 899 | eth | other | non_recyclable | 0.34 | 8.16 |
| 921 | old_town | pet | recyclable | 0.08 | 1.81 |
| 921 | old_town | metal_alu | recyclable | 0.03 | 0.70 |
| 921 | old_town | glass | recyclable | 0.30 | 6.89 |
| 921 | old_town | paper | recyclable | 0.40 | 9.32 |
| 921 | old_town | other | non_recyclable | 1.52 | 35.21 |
| 916 | old_town | pet | recyclable | 0.11 | 1.74 |
| 916 | old_town | metal_alu | recyclable | 0.04 | 0.70 |
| 916 | old_town | glass | recyclable | 0.92 | 14.63 |
| 916 | old_town | paper | recyclable | 1.01 | 16.20 |
| 916 | old_town | other | non_recyclable | 1.99 | 31.73 |
Live Coding Exercise
ae-13-data-wrangling-tidyr
- Back to
ae-13a-tidyr.qmd
Break Two

10:00
Part 3: dplyr - joining data
We…
…have multiple data frames
…want to bring them together
Data: Women in science
Information on 10 women in science who changed the world
| name |
|---|
| Ada Lovelace |
| Marie Curie |
| Janaki Ammal |
| Chien-Shiung Wu |
| Katherine Johnson |
| Rosalind Franklin |
| Vera Rubin |
| Gladys West |
| Flossie Wong-Staal |
| Jennifer Doudna |
Inputs
| name | profession |
|---|---|
| Ada Lovelace | Mathematician |
| Marie Curie | Physicist and Chemist |
| Janaki Ammal | Botanist |
| Chien-Shiung Wu | Physicist |
| Katherine Johnson | Mathematician |
| Rosalind Franklin | Chemist |
| Vera Rubin | Astronomer |
| Gladys West | Mathematician |
| Flossie Wong-Staal | Virologist and Molecular Biologist |
| Jennifer Doudna | Biochemist |
| name | birth_year | death_year |
|---|---|---|
| Janaki Ammal | 1897 | 1984 |
| Chien-Shiung Wu | 1912 | 1997 |
| Katherine Johnson | 1918 | 2020 |
| Rosalind Franklin | 1920 | 1958 |
| Vera Rubin | 1928 | 2016 |
| Gladys West | 1930 | NA |
| Flossie Wong-Staal | 1947 | NA |
| Jennifer Doudna | 1964 | NA |
| name | known_for |
|---|---|
| Ada Lovelace | first computer algorithm |
| Marie Curie | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
| Janaki Ammal | hybrid species, biodiversity protection |
| Chien-Shiung Wu | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | calculations of orbital mechanics critical to sending the first Americans into space |
| Vera Rubin | existence of dark matter |
| Gladys West | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
Desired output
| name | profession | birth_year | death_year | known_for |
|---|---|---|---|---|
| Ada Lovelace | Mathematician | NA | NA | first computer algorithm |
| Marie Curie | Physicist and Chemist | NA | NA | theory of radioactivity, discovery of elements polonium and radium, first woman to win a Nobel Prize |
| Janaki Ammal | Botanist | 1897 | 1984 | hybrid species, biodiversity protection |
| Chien-Shiung Wu | Physicist | 1912 | 1997 | confim and refine theory of radioactive beta decy, Wu experiment overturning theory of parity |
| Katherine Johnson | Mathematician | 1918 | 2020 | calculations of orbital mechanics critical to sending the first Americans into space |
| Rosalind Franklin | Chemist | 1920 | 1958 | NA |
| Vera Rubin | Astronomer | 1928 | 2016 | existence of dark matter |
| Gladys West | Mathematician | 1930 | NA | mathematical modeling of the shape of the Earth which served as the foundation of GPS technology |
| Flossie Wong-Staal | Virologist and Molecular Biologist | 1947 | NA | first scientist to clone HIV and create a map of its genes which led to a test for the virus |
| Jennifer Doudna | Biochemist | 1964 | NA | one of the primary developers of CRISPR, a ground-breaking technology for editing genomes |
Inputs, reminder
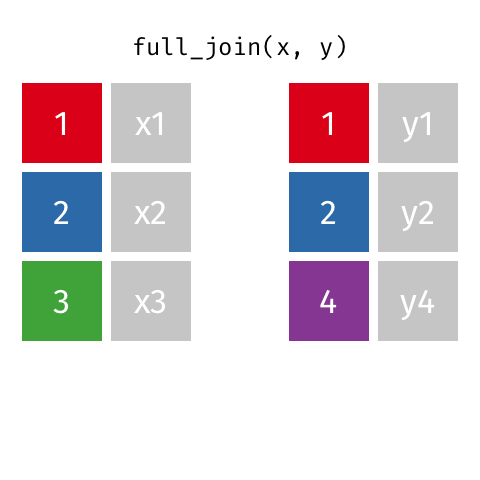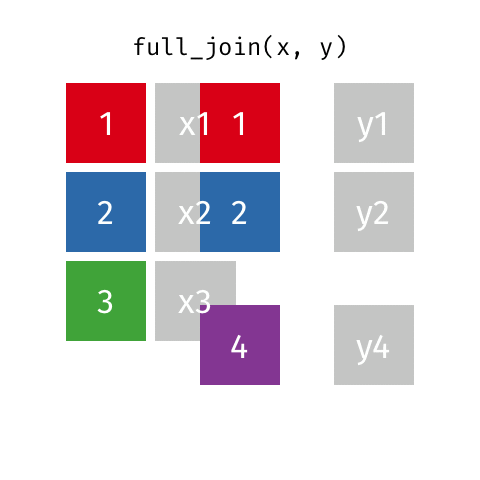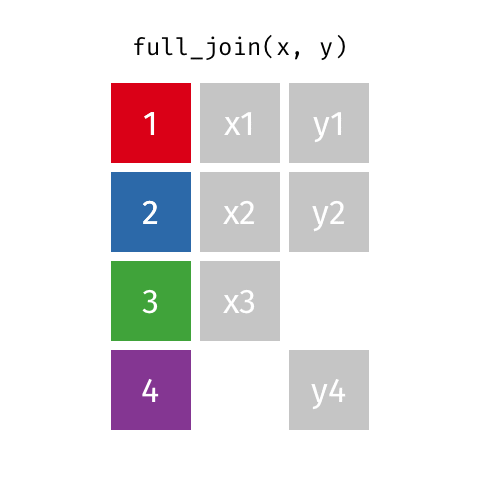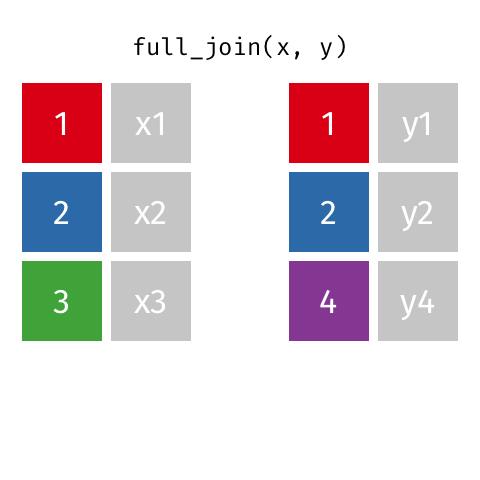
Joining data frames
Joining data frames
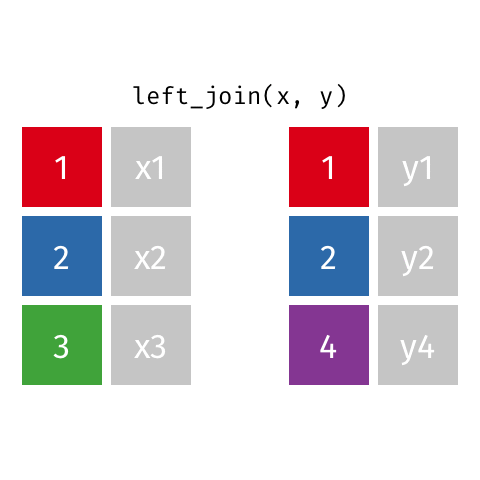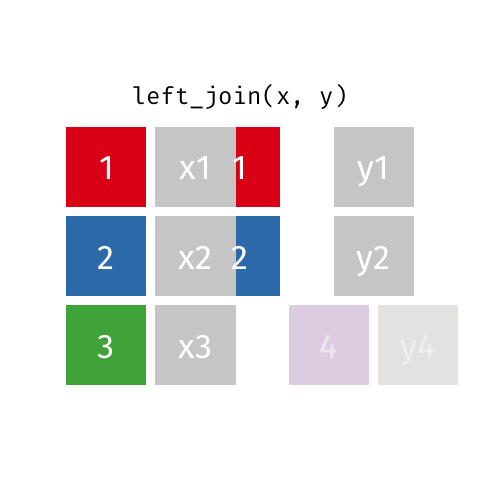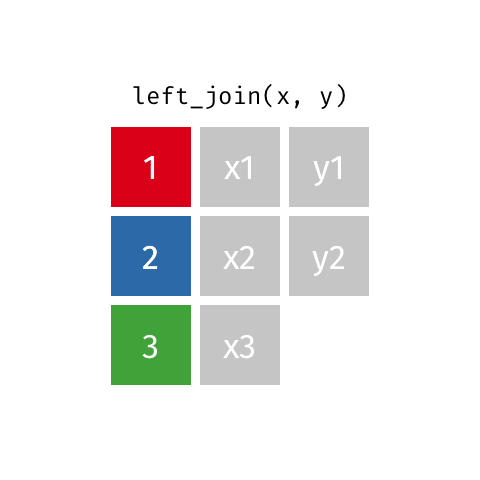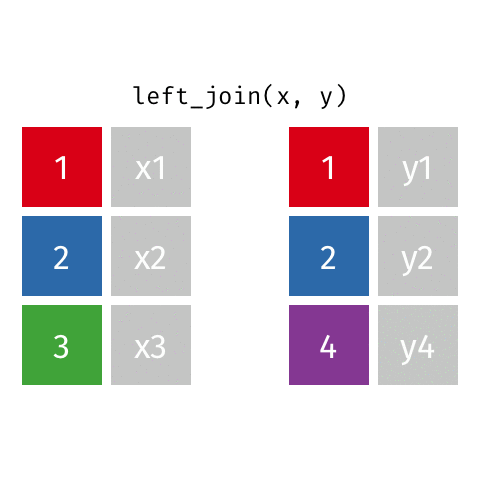
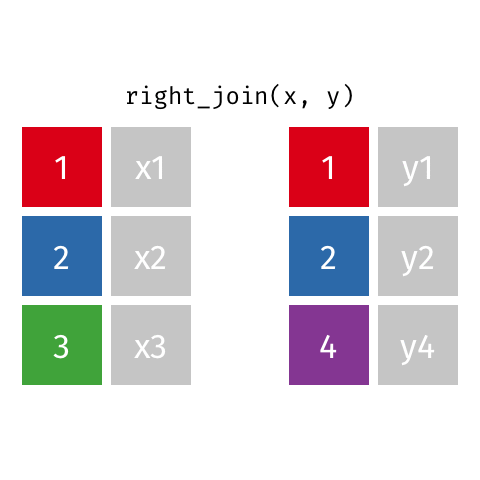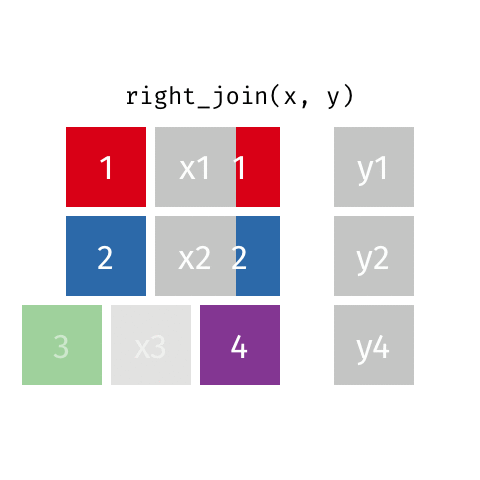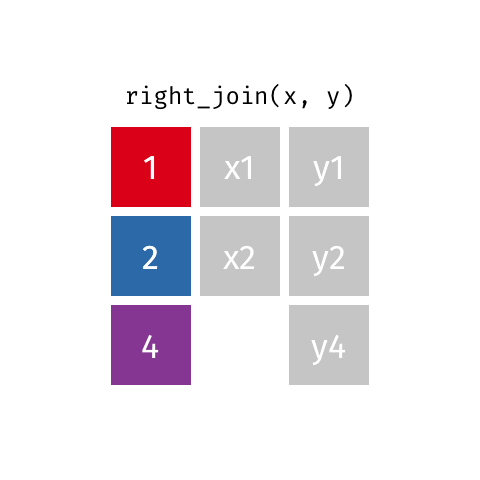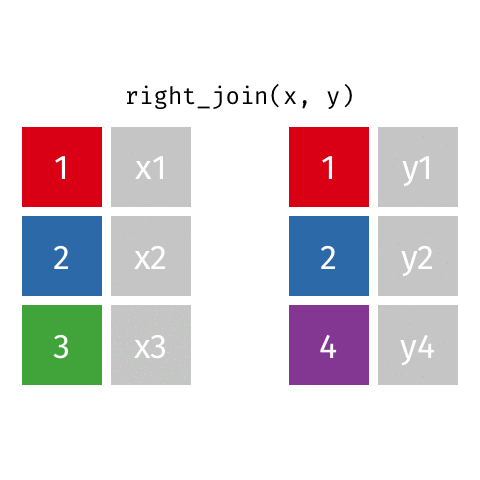
left_join(): all rows from xright_join(): all rows from yfull_join(): all rows from both x and y- …
Setup
For the next few slides…
left_join()
left_join()
# A tibble: 10 × 4
name profession birth_year death_year
<chr> <chr> <dbl> <dbl>
1 Ada Lovelace Mathematician NA NA
2 Marie Curie Physicist and Chemist NA NA
3 Janaki Ammal Botanist 1897 1984
4 Chien-Shiung Wu Physicist 1912 1997
5 Katherine Johnson Mathematician 1918 2020
6 Rosalind Franklin Chemist 1920 1958
# … with 4 more rowsright_join()
right_join()
# A tibble: 8 × 4
name profession birth_year death_year
<chr> <chr> <dbl> <dbl>
1 Janaki Ammal Botanist 1897 1984
2 Chien-Shiung Wu Physicist 1912 1997
3 Katherine Johnson Mathematician 1918 2020
4 Rosalind Franklin Chemist 1920 1958
5 Vera Rubin Astronomer 1928 2016
6 Gladys West Mathematician 1930 NA
# … with 2 more rowsfull_join()
full_join()
# A tibble: 10 × 4
name birth_year death_year known_for
<chr> <dbl> <dbl> <chr>
1 Janaki Ammal 1897 1984 hybrid species, biodiv…
2 Chien-Shiung Wu 1912 1997 confim and refine theo…
3 Katherine Johnson 1918 2020 calculations of orbita…
4 Rosalind Franklin 1920 1958 <NA>
5 Vera Rubin 1928 2016 existence of dark matt…
6 Gladys West 1930 NA mathematical modeling …
# … with 4 more rowsPutting it altogether
# A tibble: 10 × 5
name profession birth_year death_year known_for
<chr> <chr> <dbl> <dbl> <chr>
1 Ada Lovelace Mathematician NA NA first co…
2 Marie Curie Physicist an… NA NA theory o…
3 Janaki Ammal Botanist 1897 1984 hybrid s…
4 Chien-Shiung Wu Physicist 1912 1997 confim a…
5 Katherine Johnson Mathematician 1918 2020 calculat…
6 Rosalind Franklin Chemist 1920 1958 <NA>
# … with 4 more rowsLive Coding Exercise
ae-13-data-wrangling-tidyr
- Back to
ae-13a-tidyr.qmd
Homework Assignment
Submission
- All details in assignment week 13
- Due: Wednesday, 26th May at 23:59 (2 points)
Evaluation
- 5 mins
- anonymous
- after each lecture
Programming
ae-13-data-wrangling-tidyr
- Open the file:
ae-13b-dplyr.qmd - Work through the exercises
- Finalise as part of your homework
Thanks! 🌻
A large proportion of slides in this presentation are either taken from or adapted from Data Science in a Box]
Slides created via revealjs and Quarto: https://quarto.org/docs/presentations/revealjs/ Access slides as PDF on GitHub
All material is licensed under Creative Commons Attribution Share Alike 4.0 International.